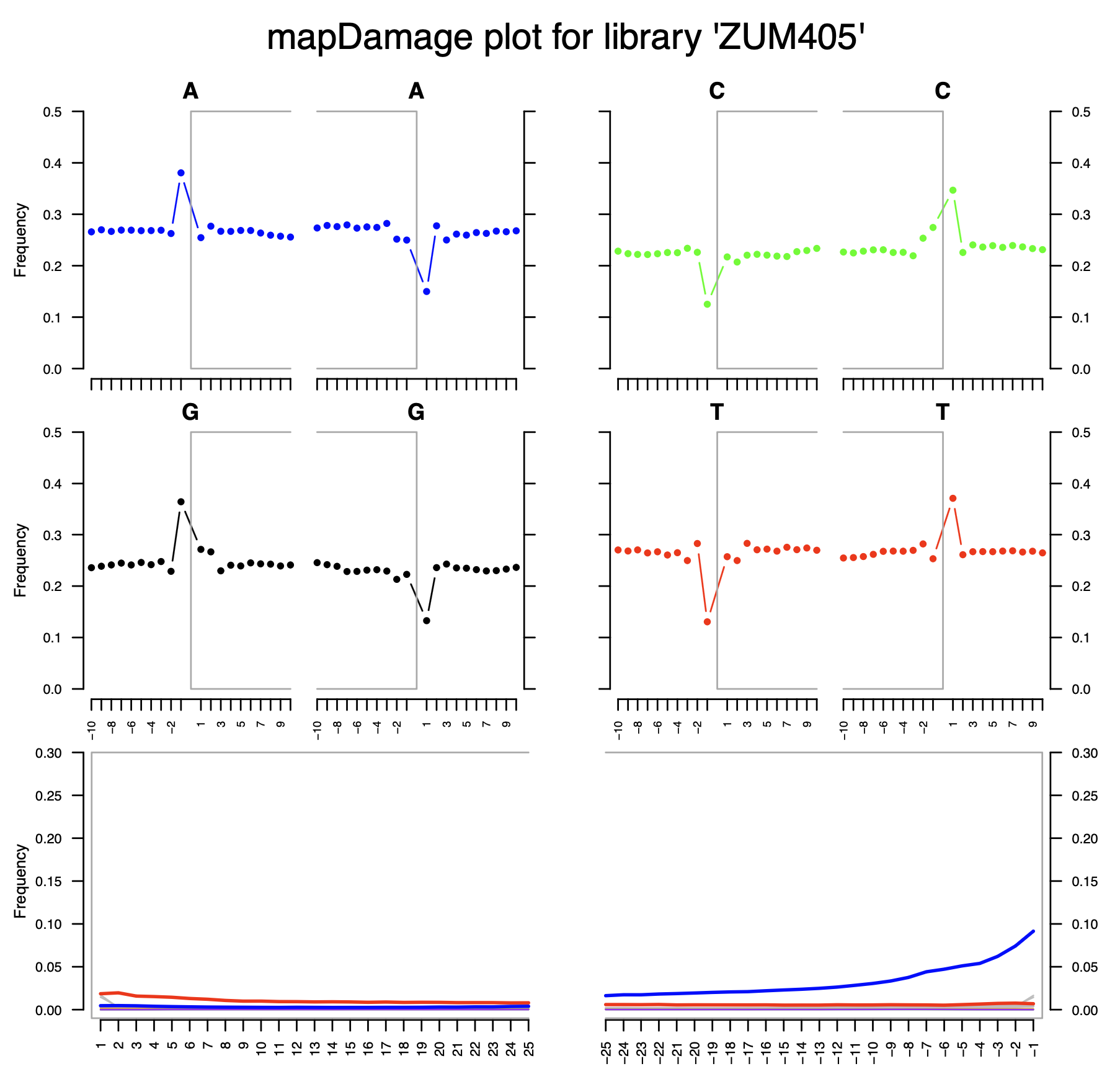
Short overview
Polar bears are a powerful symbol of the strength and endurance of the Arctic. Habitat fragmentation and contraction caused by human activities in the past century pose a serious threat to the genetic diversity of many species, including polar bears, and increase their risk of extinction. The aim of this study is to investigate the genetic diversity, population structure, migration trends, and demographic history of polar bear populations in the Russian Arctic.
Extended project overview
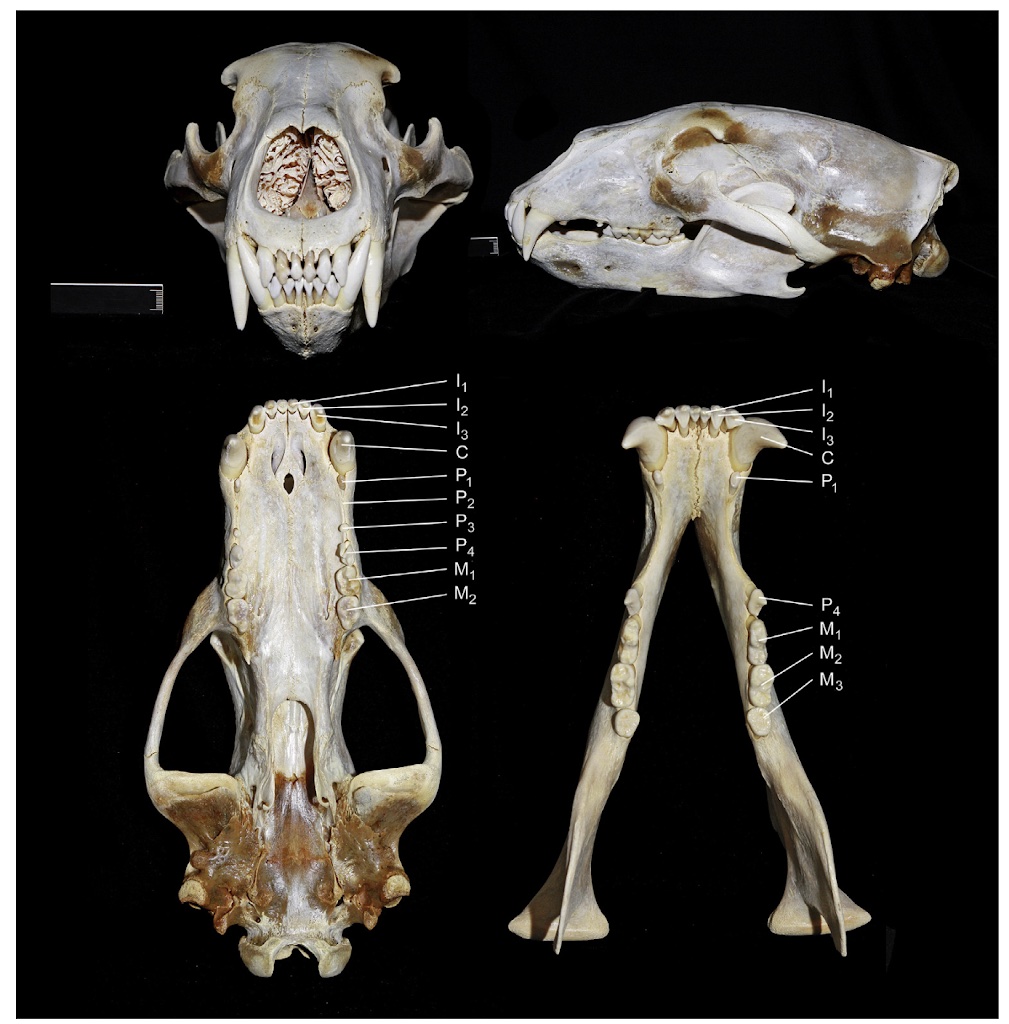
The polar bear is the world’s largest terrestrial carnivore and a dominant predator in the Arctic, roaming the sea ice and tundra. This charismatic species is the only land mammal that uses pack ice as its main habitat. The polar bear’s Latin name, Ursus maritimus, means “sea bear.”. These bears are strong swimmers and can cover vast distances in icy waters.
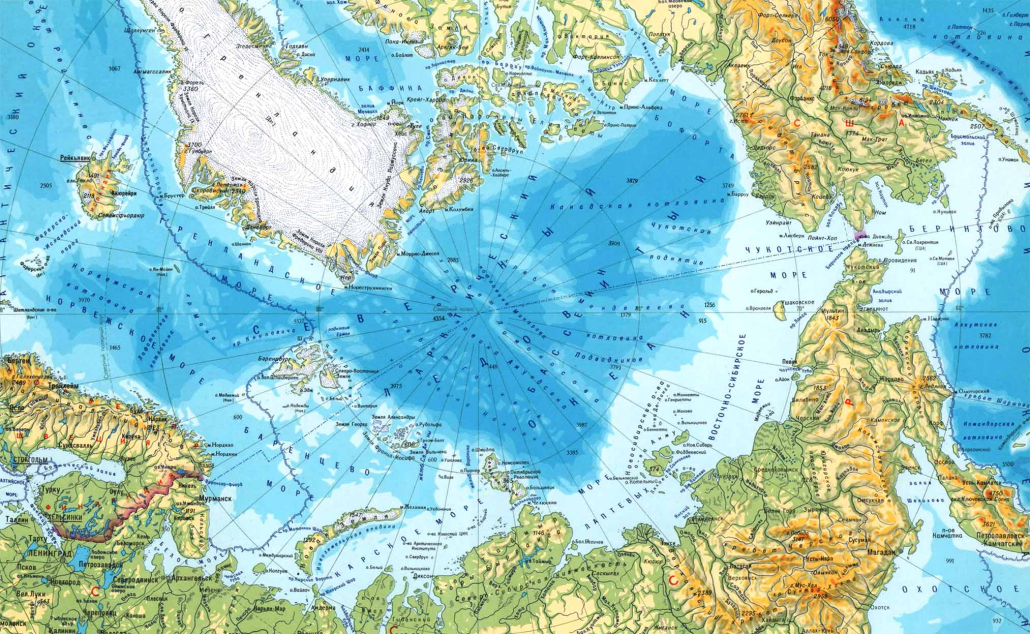
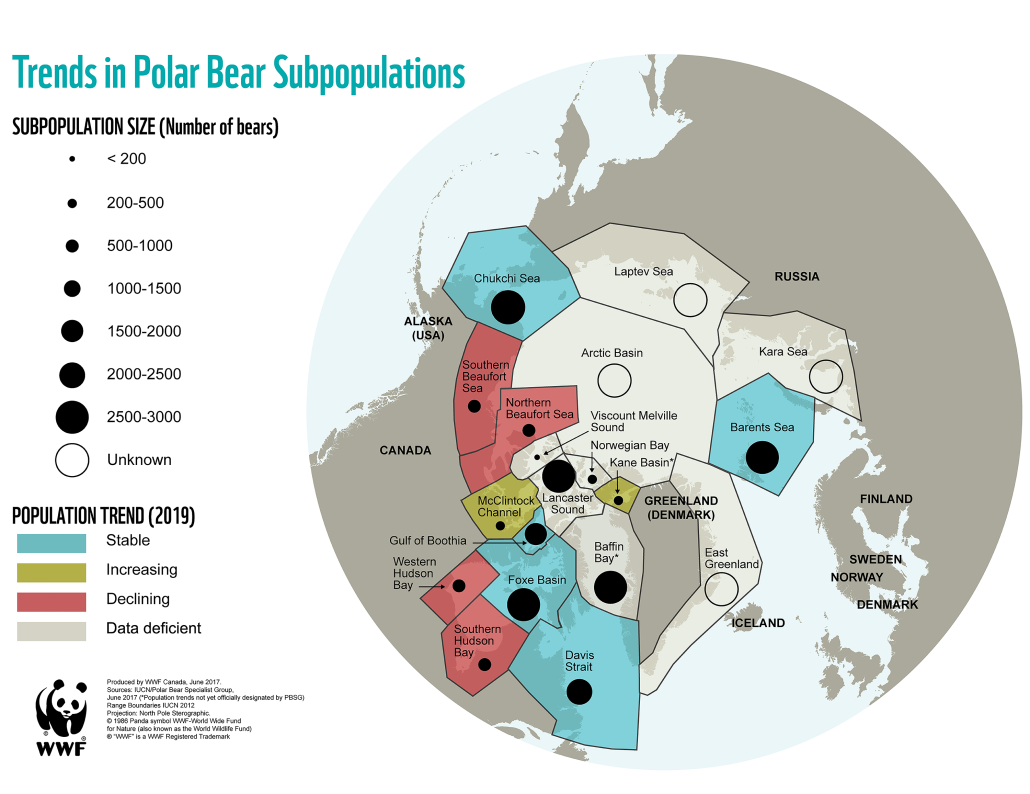
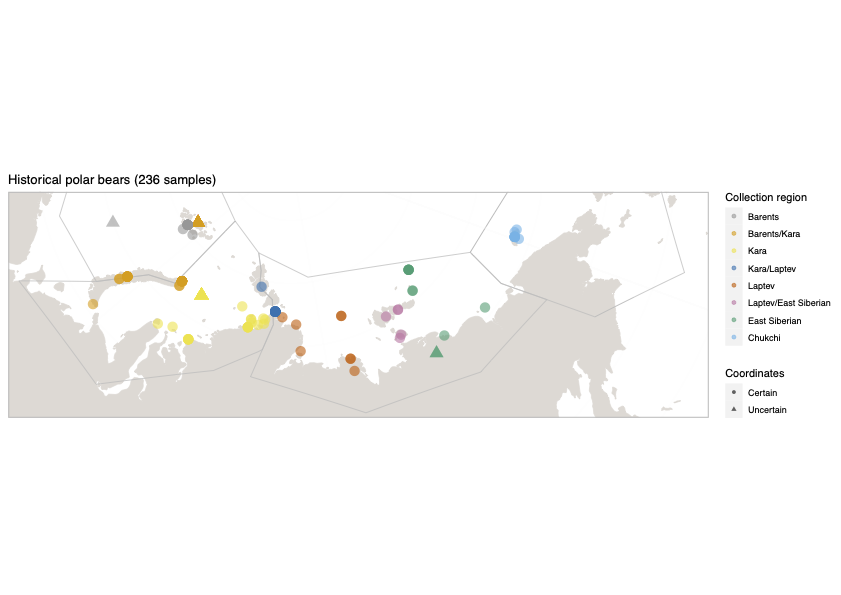
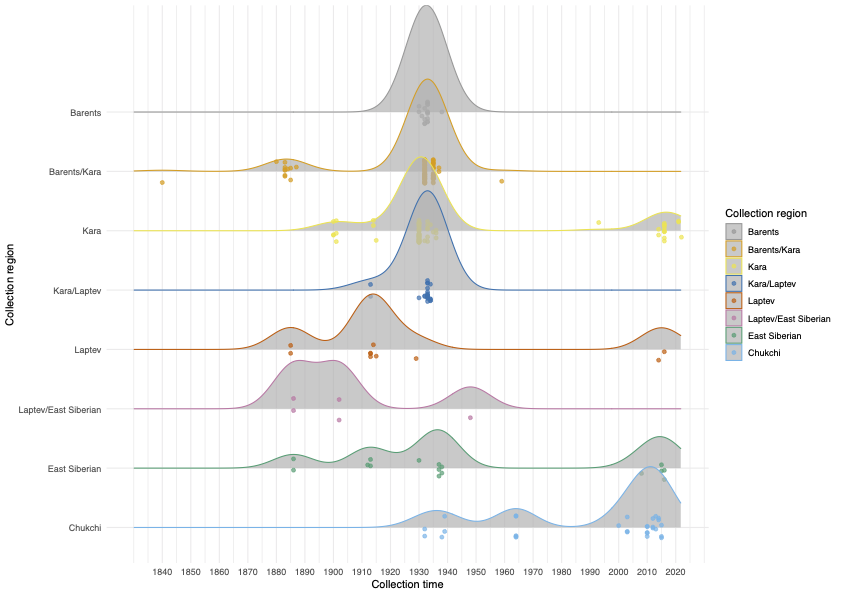
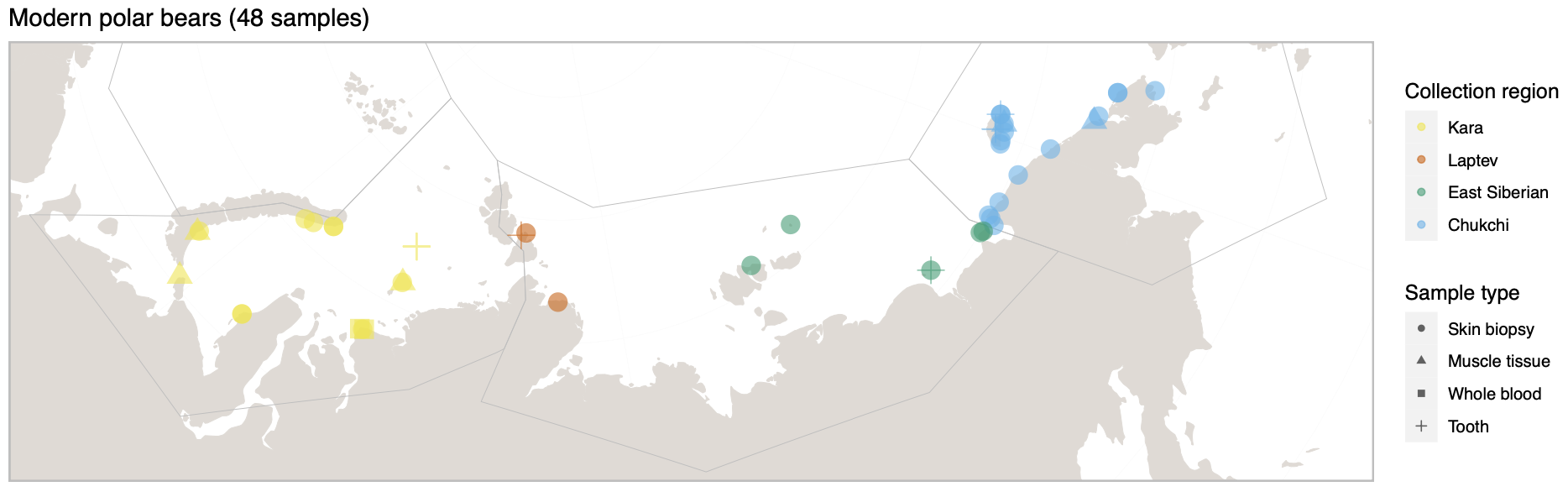
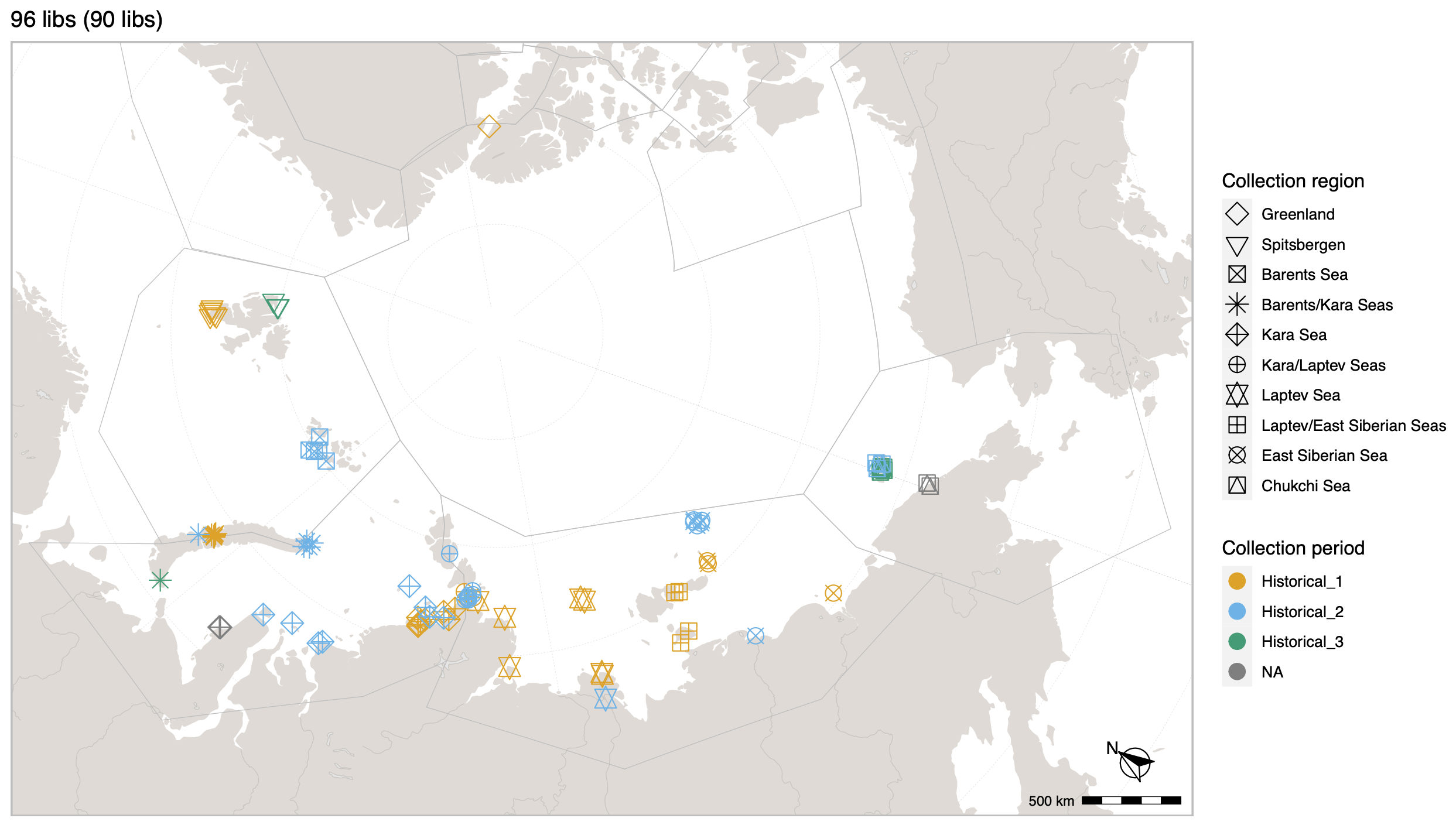
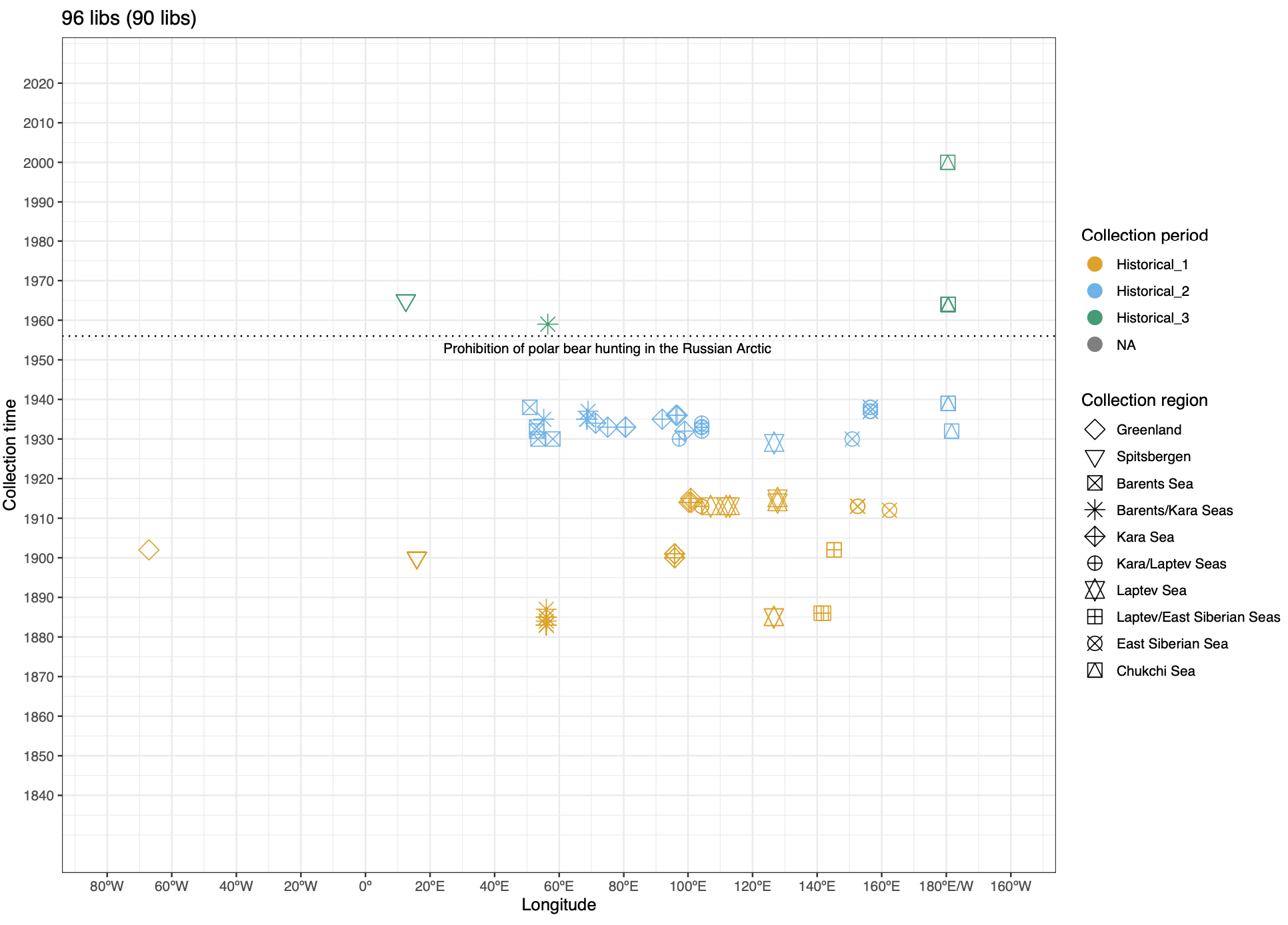
Polar bears are distributed in 20 relatively distinct populations across the circumpolar Arctic, ranging in size from a few hundred to a few thousand individuals. In the Russian Arctic, there are three subpopulations: the Kara-Barents, Laptev, and Chukotka-Alaska populations. However, the genetic status of these subpopulations is largely unknown, which limits our conservation efforts and ability to protect polar bears.
The past century has seen an increase in human activity and infrastructure development in the Russian Arctic, leading to animal habitat fragmentation and contraction. This poses a significant threat to the genetic diversity of many species, including polar bears, and increases their risk of extinction.
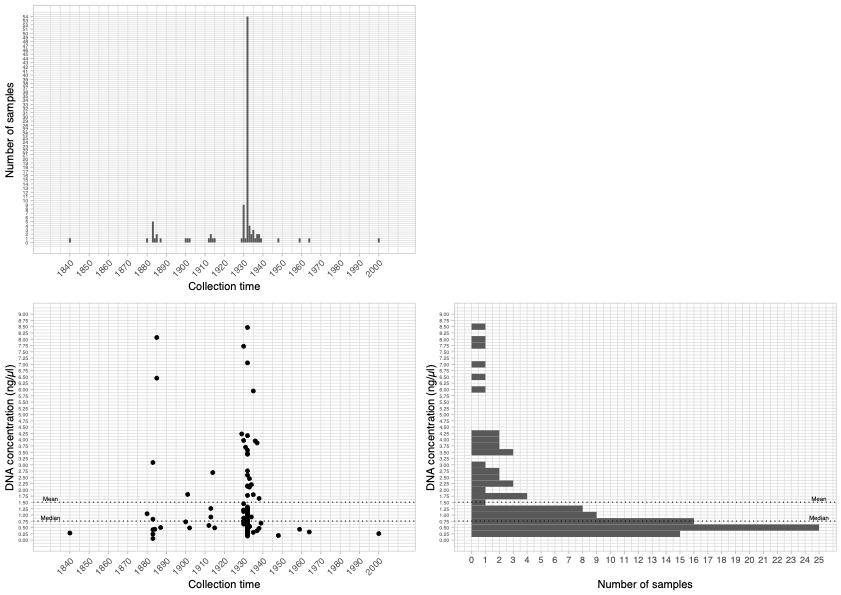
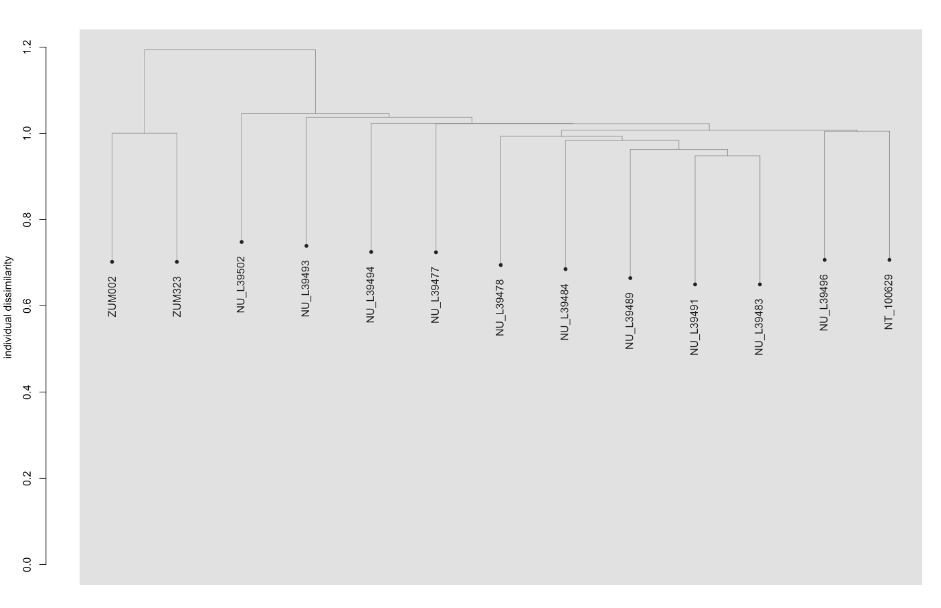
To address these issues, this project aims to uncover the genetic diversity, population structure, and demographic history of polar bear subpopulations in the Russian Arctic. Specifically, the study seeks to answer the question: “How has the genetic status of polar bear subpopulations changed since the 1900s?”

By providing this information, we can better understand the threats facing polar bears and take more effective action to protect their well-being.
 © Hans-Jurgen Mager
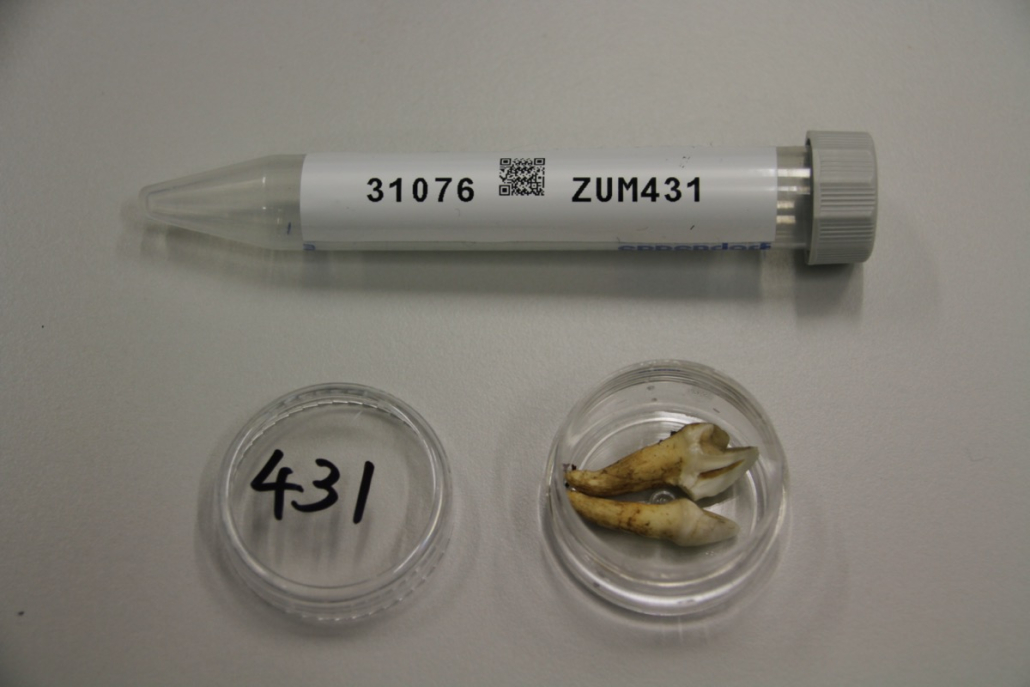
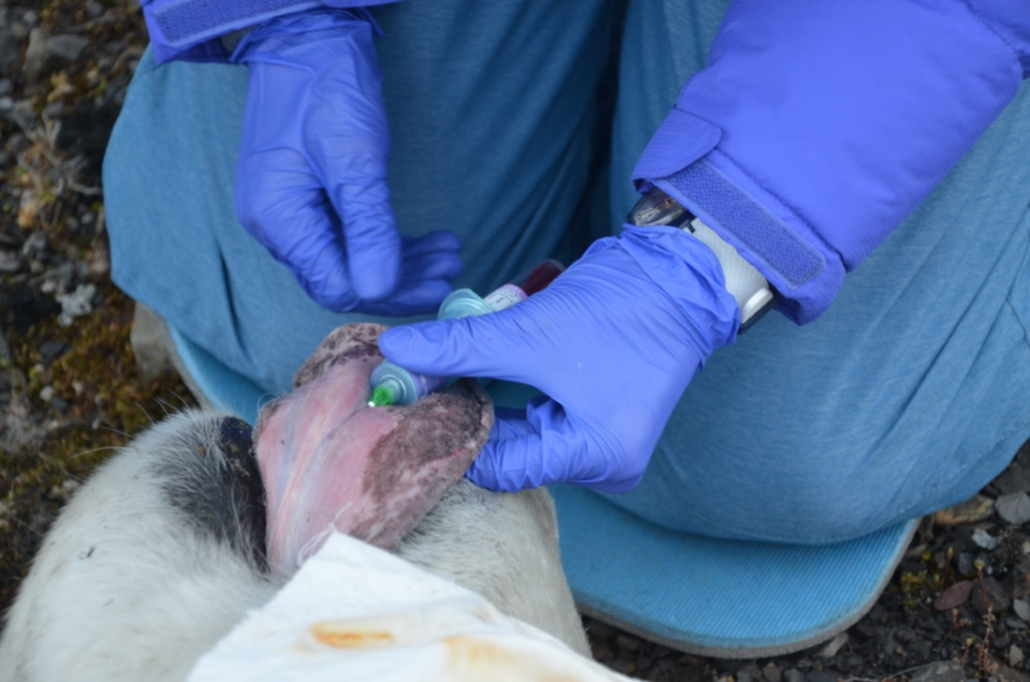
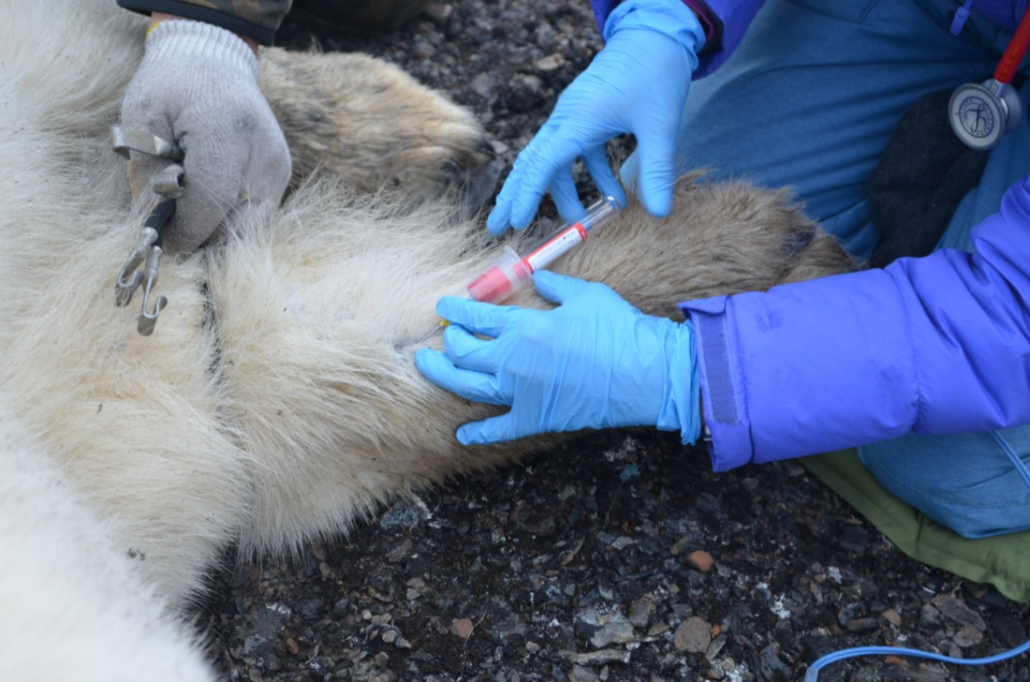
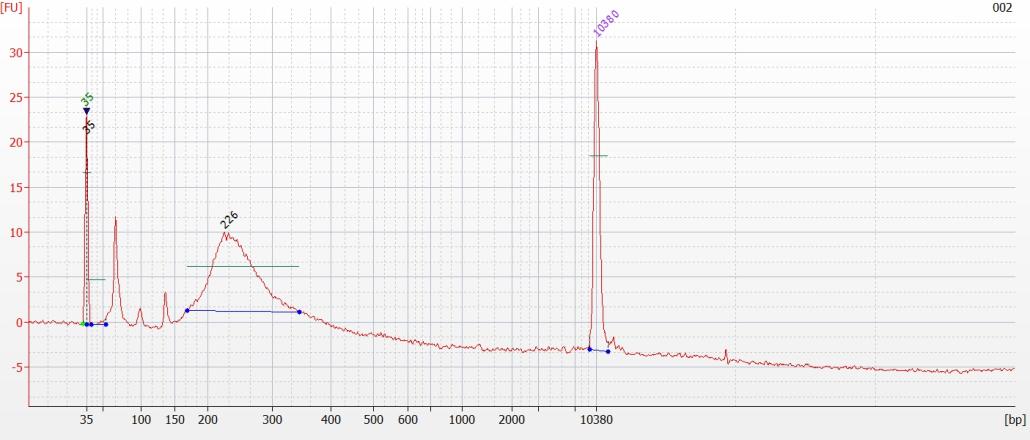
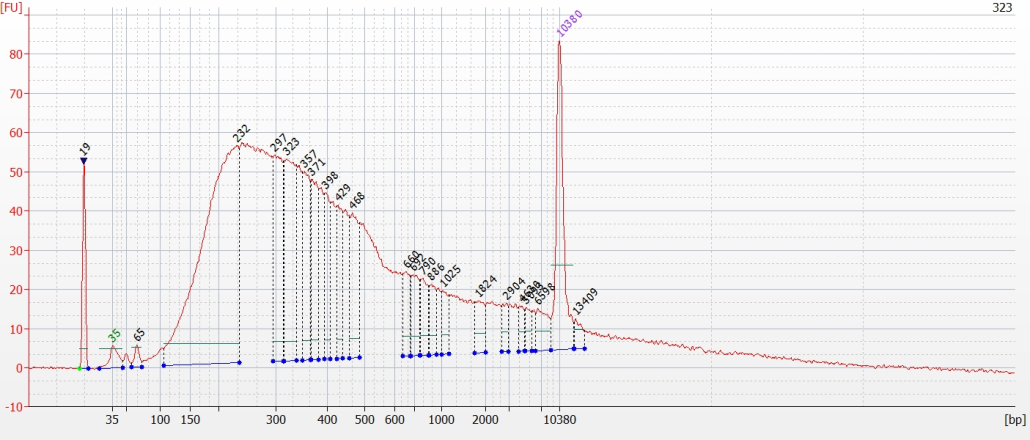
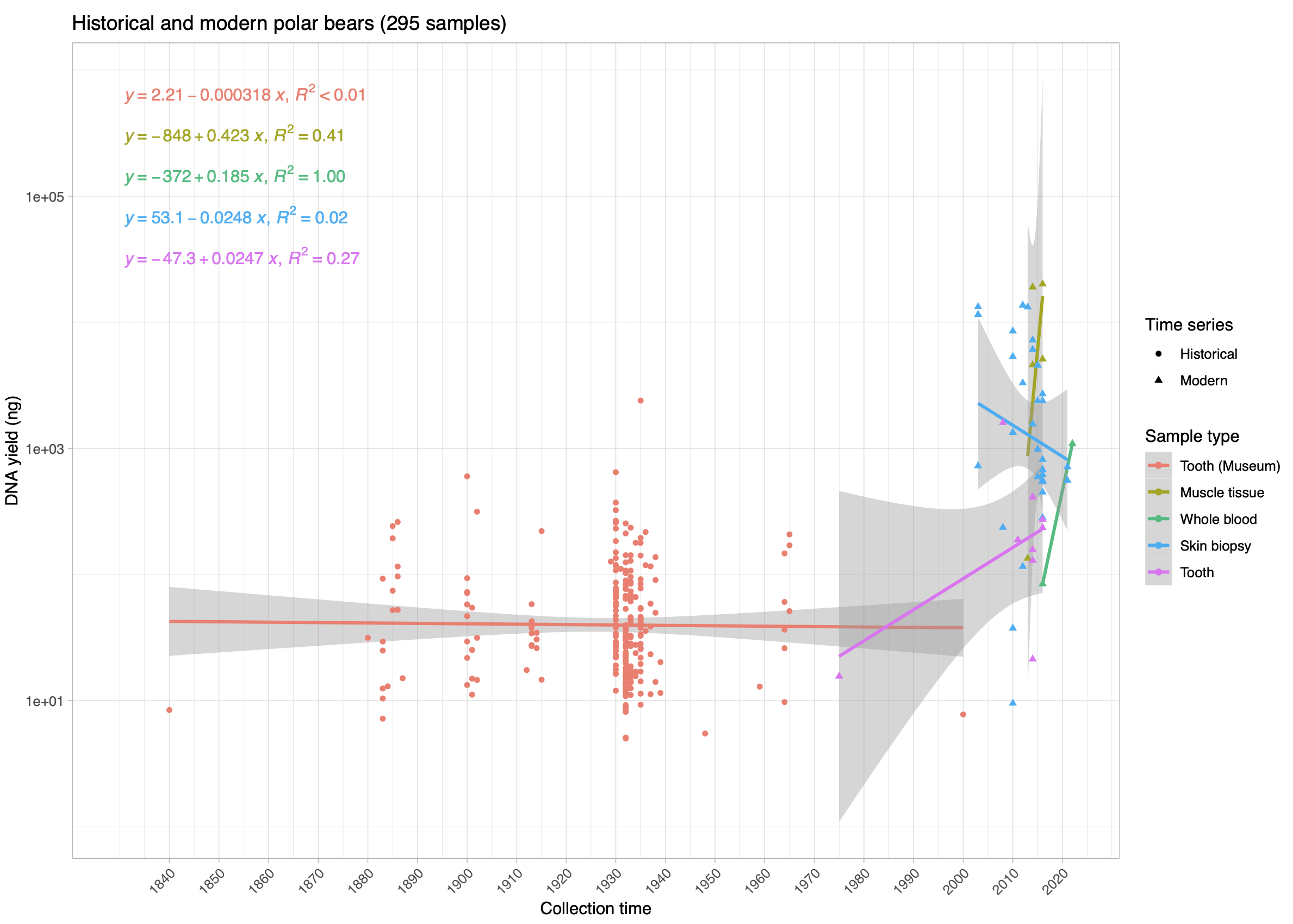
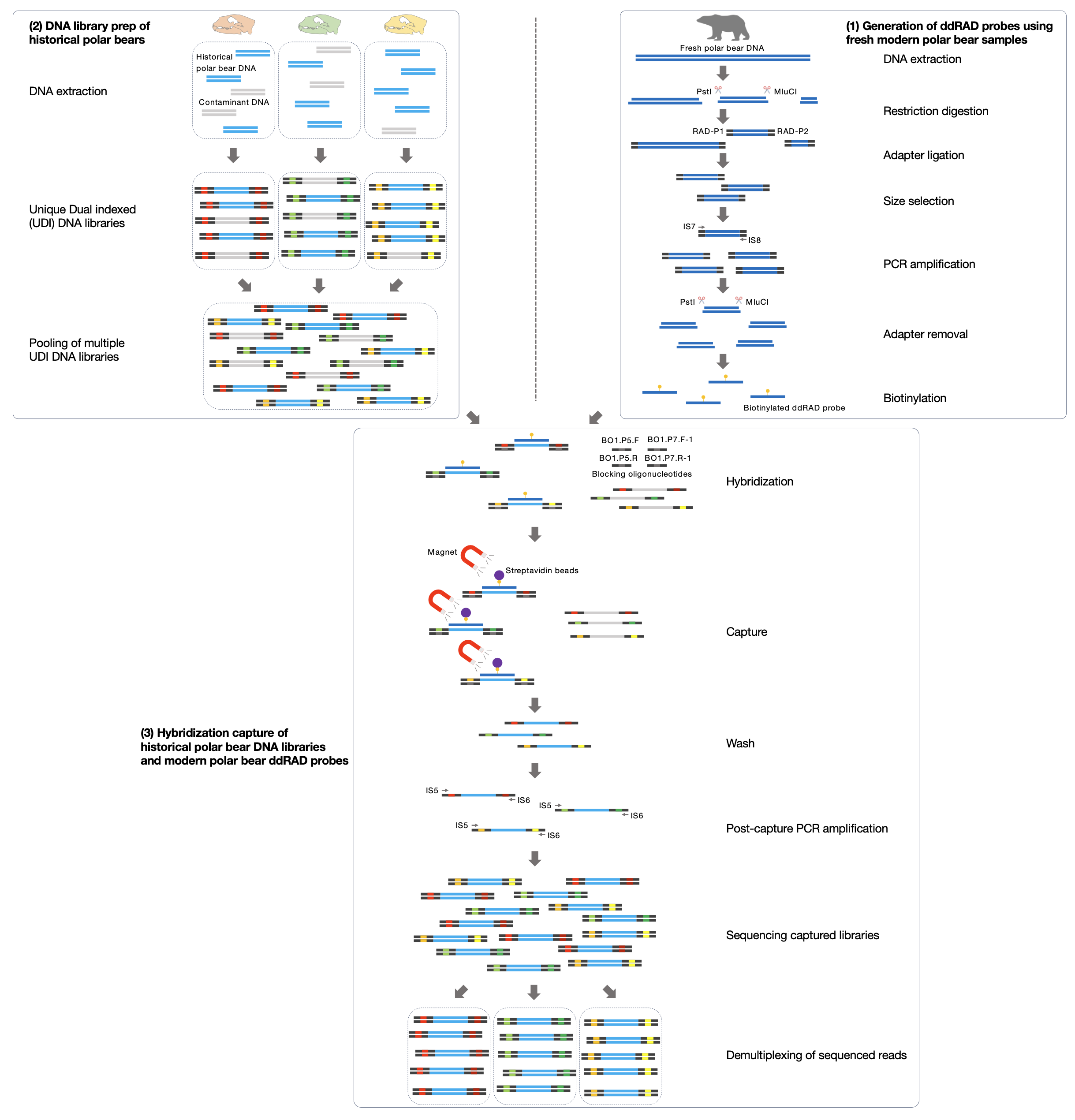
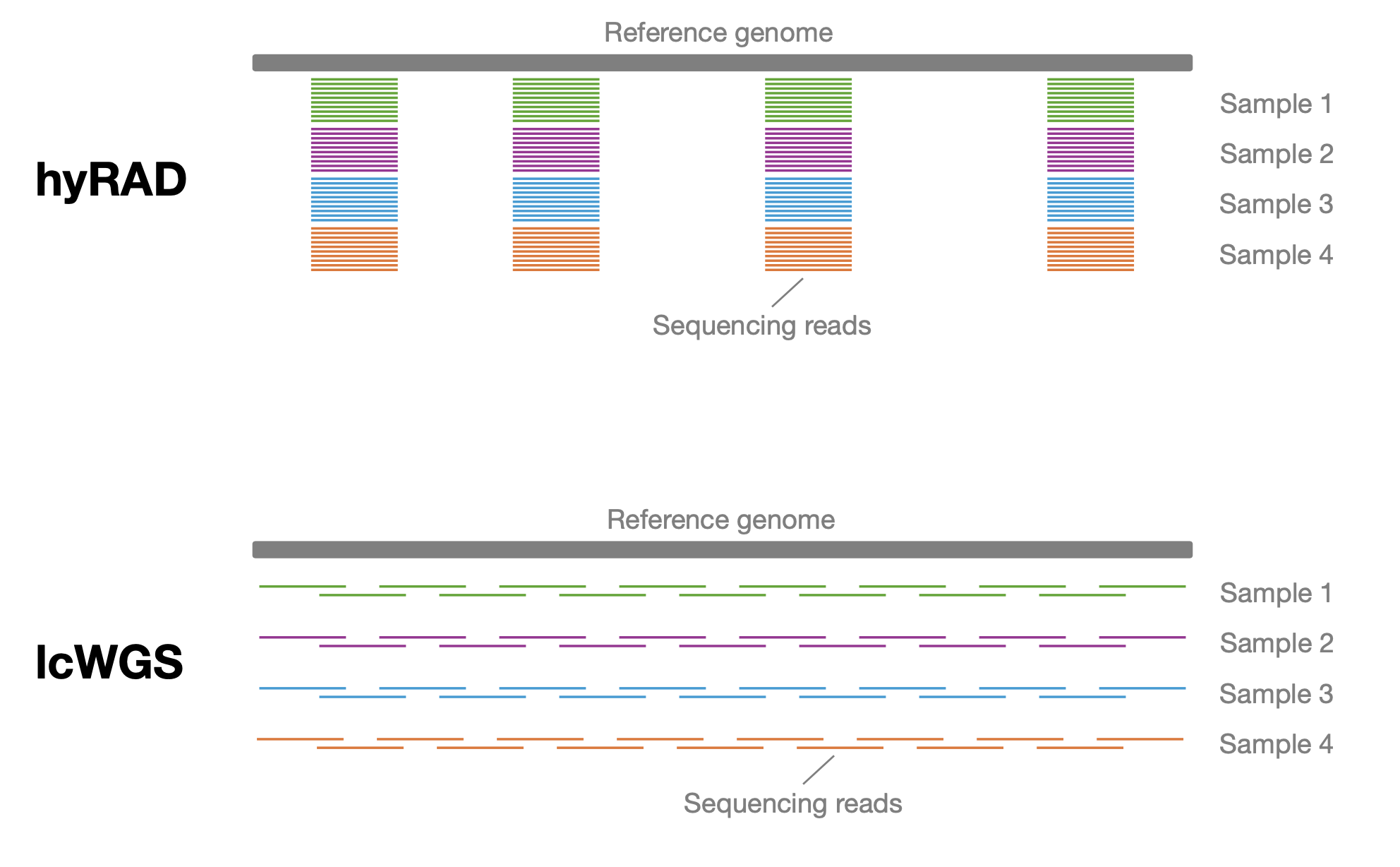
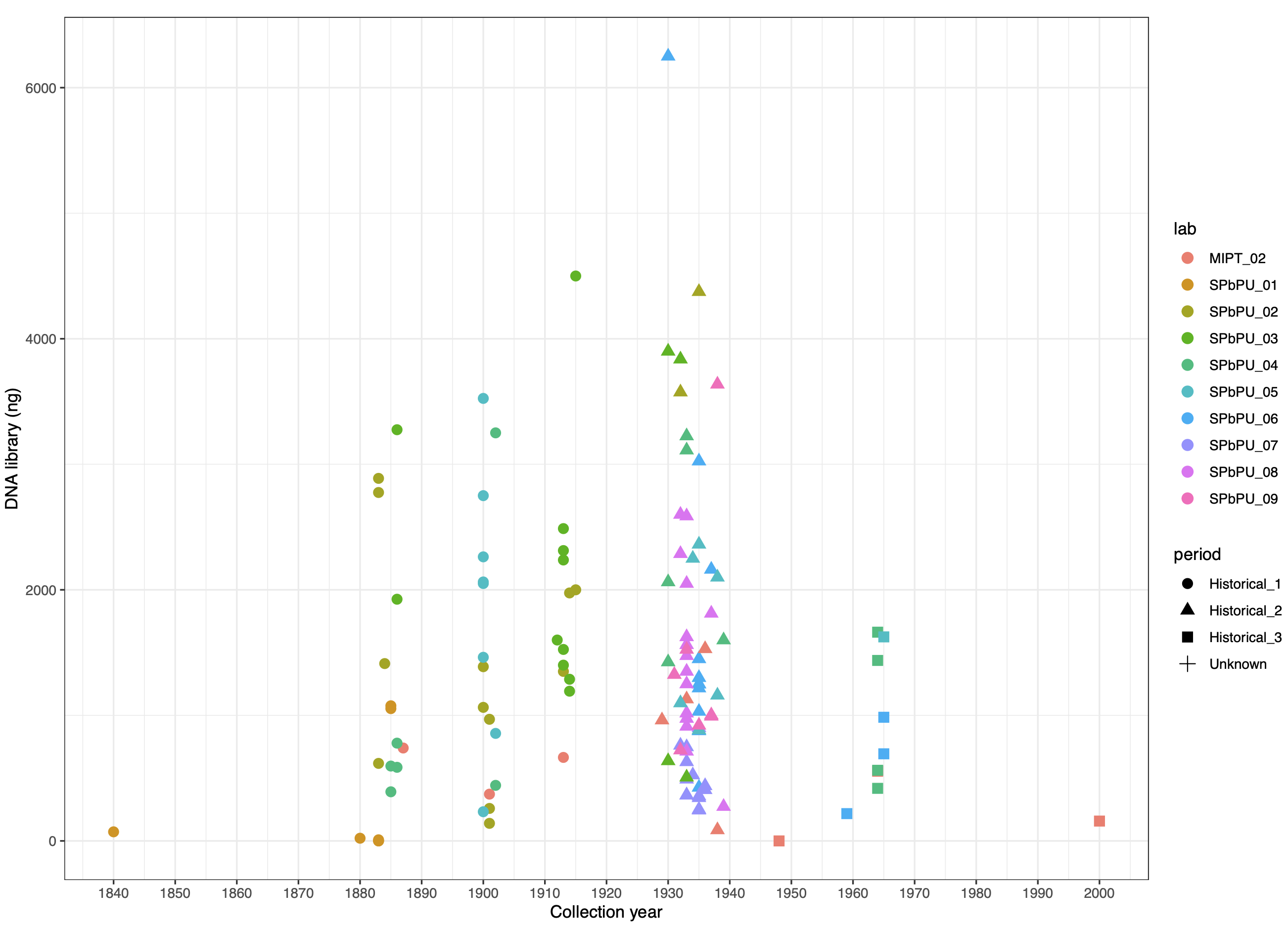
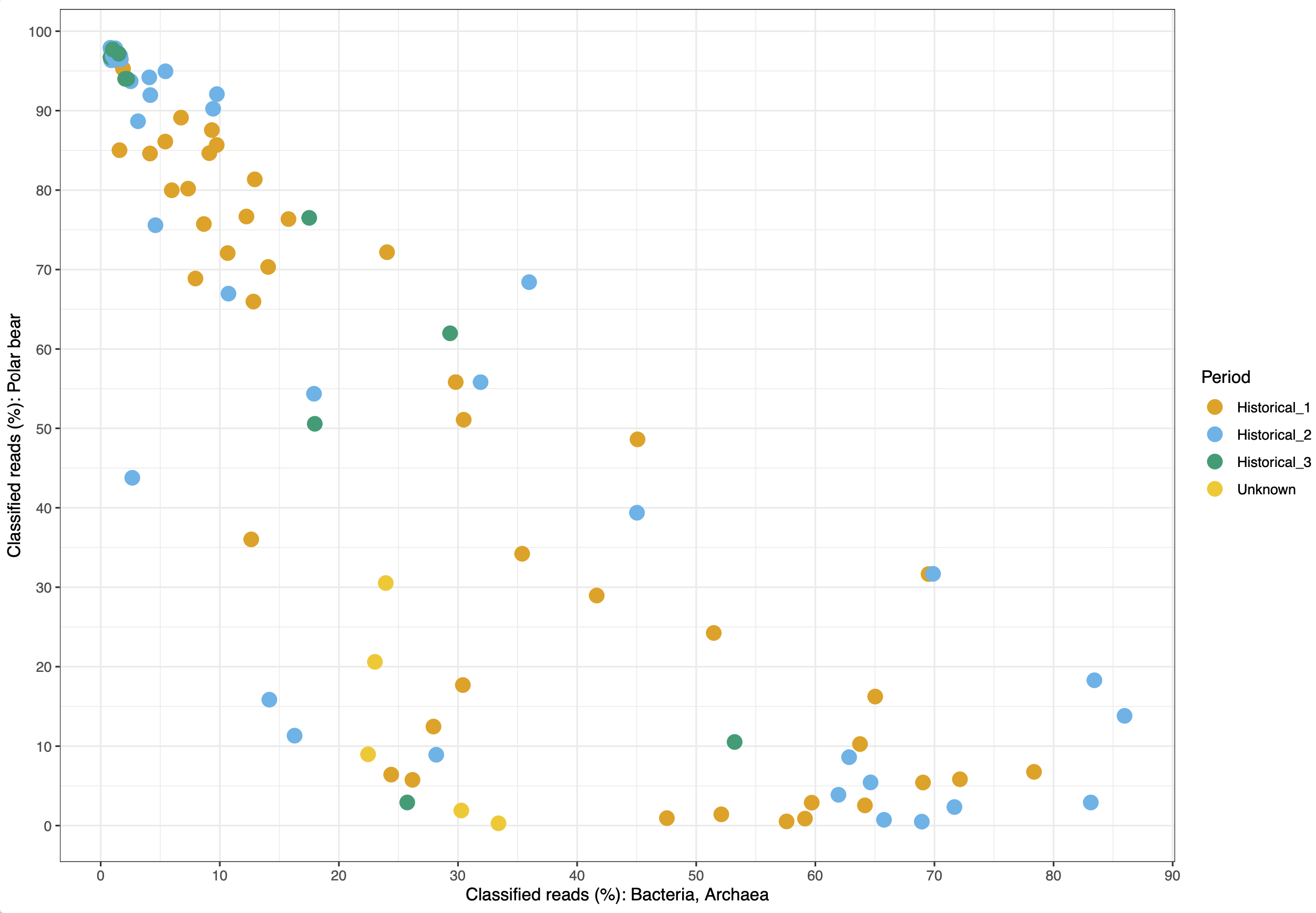
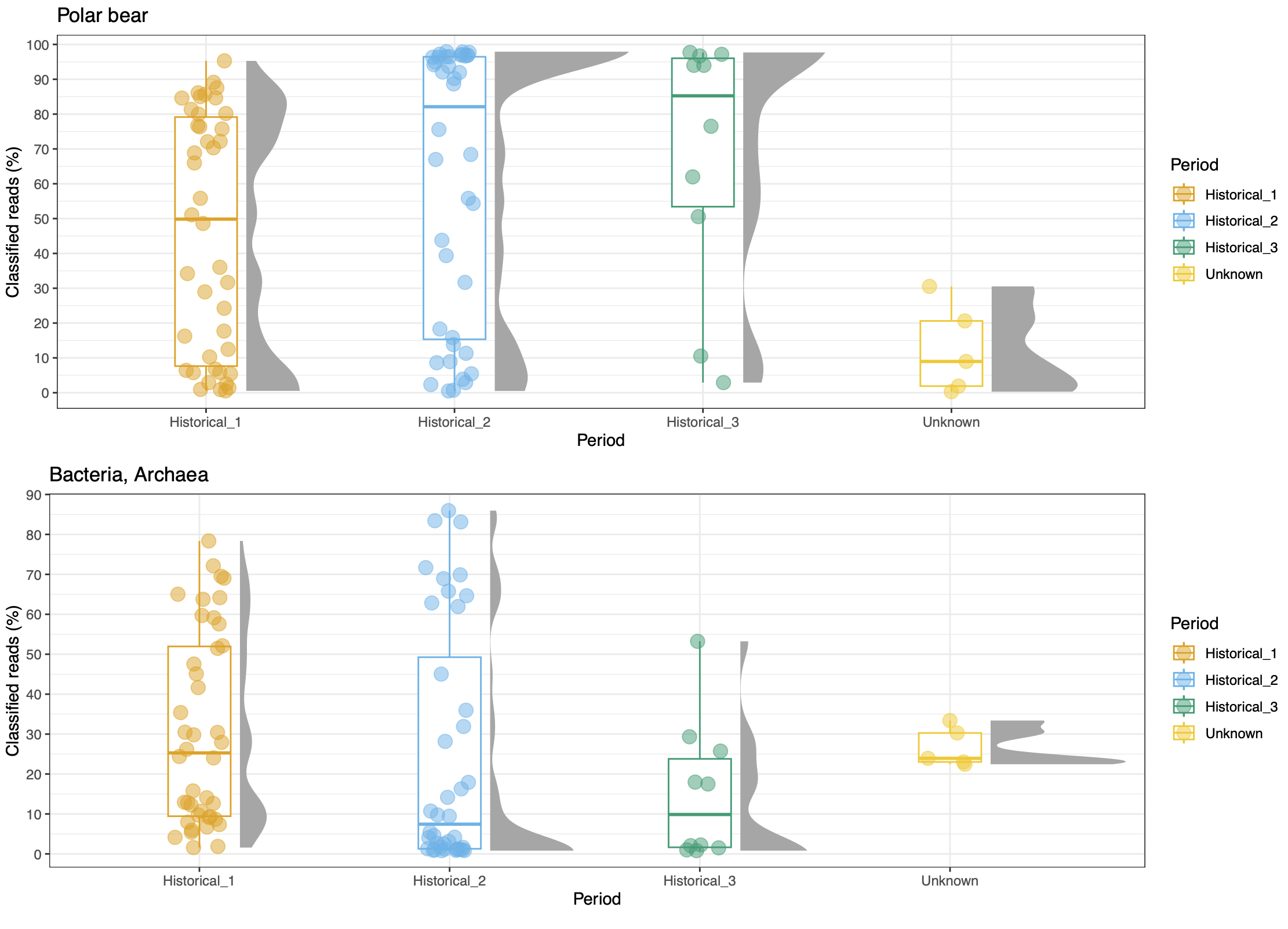
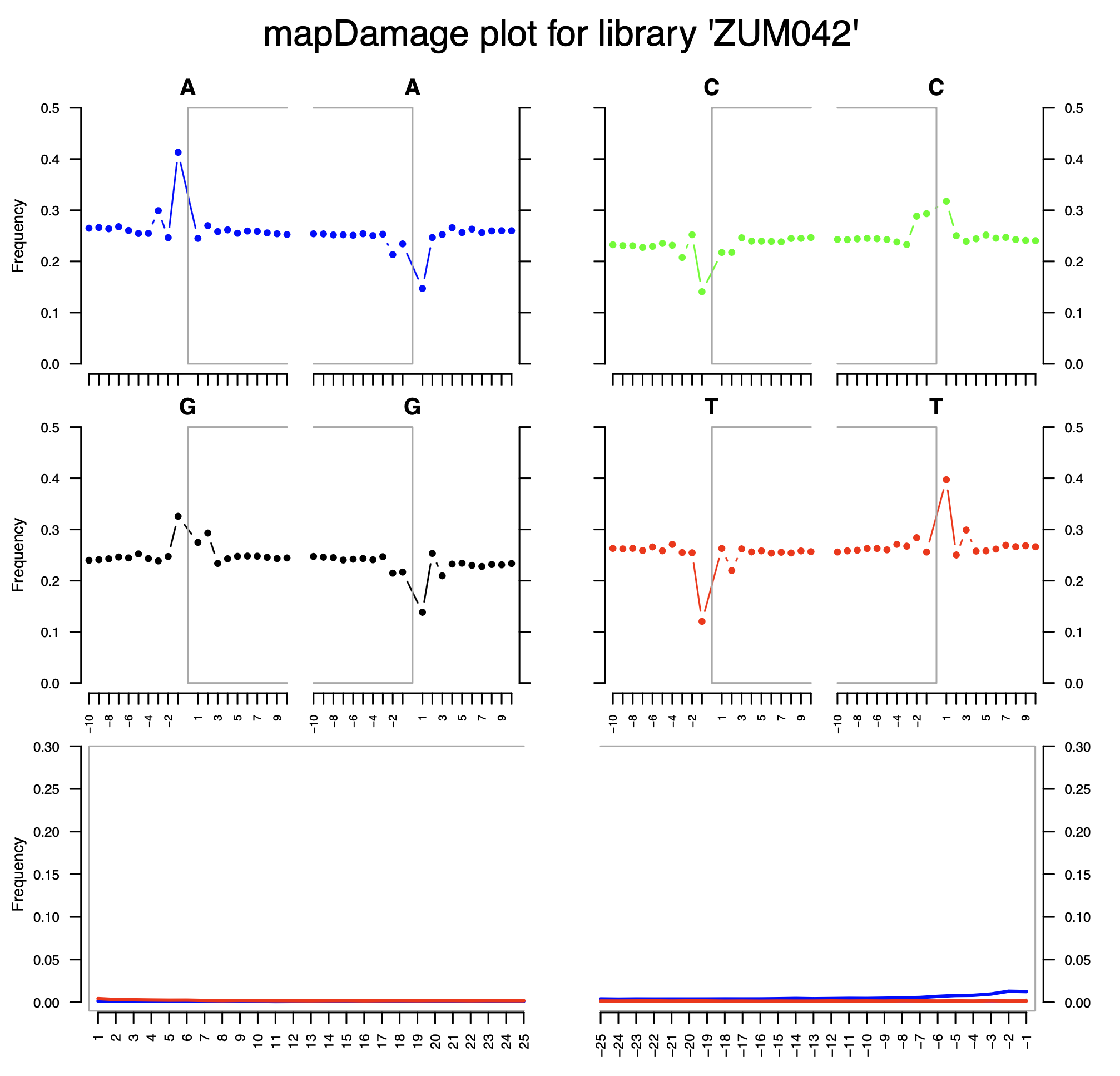
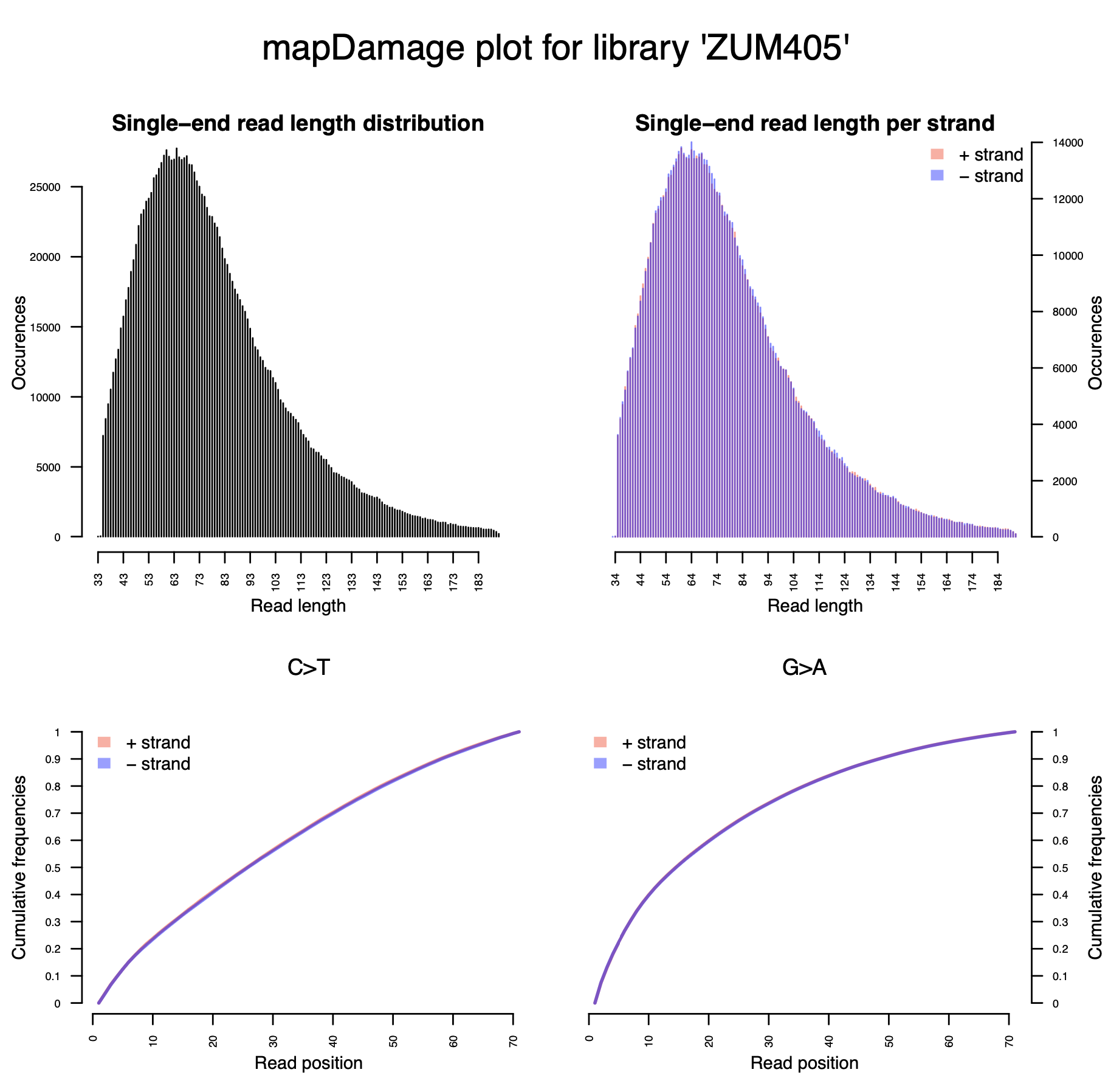
© Hans-Jurgen MagerThis project aims to provide a comprehensive understanding of the genetic variability, population structure, and demographic history of polar bears in the Russian Arctic. By sequencing archival and modern DNA samples collected from various habitats in the region, including samples dating back to the 1880s, we aim to produce a detailed description of the polar bear population in the Russian Arctic. The project’s results have the potential to fill key gaps in our knowledge of polar bear population genetics and help us understand the impact of human activity on polar bear populations in the past century. Additionally, the findings will facilitate the development of molecular genetic tools for preventing poaching and monitoring migrations.
Overall, this project is an essential step towards protecting the well-being of polar bears and preserving the fragile Arctic ecosystem in which they live.
 © Dan Bolton
© Dan Bolton